Abstract
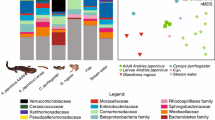
Pathogens currently threaten the existence of many amphibian species. In efforts to combat global declines, researchers have characterized the amphibian cutaneous microbiome as a resource for disease management. Characterization of microbial communities has become useful in studying the links between organismal health and the host microbiome. Hellbender salamanders (Cryptobranchus alleganiensis) provide an ideal system to explore the cutaneous microbiome as this species requires extensive conservation management across its range. In addition, the Ozark hellbender subspecies (Cryptobranchus alleganiensis bishopi) exhibits chronic wounds hypothesized to be caused by bacterial infections, whereas the eastern hellbender (Cryptobranchus alleganiensis alleganiensis) does not. We assessed the cutaneous bacterial microbiome of both subspecies at two locations in the state of Missouri, USA. Through 16S rRNA gene-based amplicon sequencing, we detected more than 1000 distinct operational taxonomic units (OTUs) in the cutaneous and environmental bacterial microbiome. Phylogenetic and abundance-based dissimilarity matrices identified differences in the bacterial communities between the two subspecies, but only the abundance-based dissimilarity matrix identified differences between wounds and healthy skin on Ozark hellbenders. The higher abundance of OTUs on Ozark wounds suggests that commensal bacteria present on the skin and environment may be opportunistically colonizing the wounds. This brief exploration of the hellbender cutaneous bacterial microbiome provides foundational support for future studies seeking to understand the hellbender cutaneous bacterial microbiome and the role of the bacterial microbiota on chronic wounds of Ozark hellbenders.




Similar content being viewed by others
References
Federici E, Rossi R, Fidati L, Paracucchi R, Scargetta S, Montalbani E, Franzetti A, La Porta G, Fagotti A, Simonceli F, Cenci G, Di Rosa I (2015) Characterization of the skin microbiota in Italian stream frogs (Rana italica) infected and uninfected by a cutaneous parasitic disease. Microbes Environ 30:262–269
Loudon AH, Holland JA, Umile TP, Burzynski EA, Minbiole KPC, Harris RN (2014) Interactions between amphibians’ symbiotic bacteria cause the production of emergent anti-fungal metabolites. Front Microbiol 5:441
Vredenburg VT, Briggs CJ, Harris RN (2011) Host-pathogen dynamics of amphibian chytridiomycosis: the role of the skin microbiome in health and disease. Fungal diseases: an emerging challenge to human, animal, and plant health. National Academic Press, Washington D.C., pp 342–355
Barker CJ, Gillett A, Polkinghorne A, Timms P (2013) Investigation of the koala (Phascolarctos cinereus) hindgut microbiome via 16S pyrosequencing. Vet Microbiol 167:554–564
Kueneman JG, Parfrey LW, Woodhams DC, Archer HM, Knight R, McKenzie VJ (2014) The amphibian skin-associated microbiome across species, space and life history stages. Mol Ecol 23:1238–1250
Shabbir MZ, Park J, Muhammad K, Rabbani M, Rana MY, Harvill ET (2014) Culture independent analysis of respiratory microbiome of houbara bustard (Chlamydotis undulata) revealed organisms of public health significance. Int J Agric Biol 16:222–226
Ishak HD, Plowes R, Sen R, Kellner K, Meyer E, Estrada DA, Dowd SE, Mueller UG (2011) Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb Ecol 61:821–831
Daszak P, Cunningham AA, Hyatt AD (2003) Infectious disease and amphibian population declines. Divers Distrib 9:141–150
Nickerson MA, Mays CE (1973) The hellbenders: North American ‘giant salamanders’. Milwaukee Public Museum, Milwaukee
Foster RL, McMillan AM, Roblee KJ (2009) Population status of hellbender salamanders (Cryptobranchus alleganiensis) in the Allegheny River drainage of New York State. J Herpetol 43:579–588
Hecht-Kardasz KA, Nickerson MA, Freake M, Colclough P (2012) Population structure of the hellbender (Cryptobranchus alleganiensis) in a Great Smoky Mountains stream. Bull Fla Museum Nat Hist 51:227
Briggler J, Utrup J, Davidson C, Humphries J, Groves J, Johnson T, Ettling J, Wanner M, Traylor-Holzer K, Reed D, Lindgren V, Byers O (2007) Hellbender population and habitat viability assessment: final report. IUCN/SSC Conservation Breeding Specialist Group, Apple Valley
Burgmeier NG, Unger SD, Sutton TM, Williams RN (2011) Population status of the eastern hellbender (Cryptobranchus alleganiensis alleganiensis) in Indiana. J Herpetol 45:195–201
Federal Register (2011) Endangered and threatened wildlife and plants; endangered status for the Ozark hellbender salamander. Fed Commun Comm 76:61956–61978
Wheeler BA, Prosen E, Mathis A, Wilkinson RF (2003) Population declines of a long-lived salamander: a 20+-year study of hellbenders, Cryptobranchus alleganiensis. Biol Conserv 109:151–156
Hiler WR, Wheeler BA, Trauth SE (2005) Abnormalities in the Ozark hellbender (Cryptobranchus alleganiensis bishopi) in Arkansas: a comparison between two rivers with a historical perspective. J Arkansas Acad Sci 59:88–94
Nickerson CA, Ott CM, Castro SL, Garcia VM, Molina TC, Briggler JT, Pitt AL, Tavano JJ, Byram JK, Barrila J et al (2011) Evaluation of microorganisms cultured from injured and repressed tissue regeneration sites in endangered giant aquatic Ozark hellbender salamanders. PLoS One 6, e28906
Wheeler BA, McCallum ML, Trauth SE (2002) Abnormalities in the Ozark hellbender, Cryptobranchus alleganiensis bishopi. J Arkansas Acad Sci 56:250–252
Irwin K (2008) Ozark hellbender long-term monitoring SWG project. Arkansas Game and Fish Commission, Benton
Hill GT, Mitkowski NA, Aldrich-Wolfe L, Emele LR, Jurkonie DD, Ficke A, Maldonado-Ramirez S, Lynch ST, Nelson EB (2000) Methods for assessing the composition and diversity of soil microbial communities. Appl Soil Ecol 15:25–36
Pace NR (1997) A molecular view of microbial diversity and the biosphere. Science 276:734–740
Jani AJ, Briggs CJ (2014) The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. Proc Natl Acad Sci U S A 111:E5049–E5058
Fitzpatrick BM, Allison AL (2014) Similarity and differentiation between bacteria associated with skin of salamanders (Plethodon jordani) and free-living assemblages. FEMS Microbiol Ecol 88:482–494
Loudon AH, Woodhams DC, Parfrey LW, Archer H, Knight R, McKenzie V, Harris RN (2014) Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus). ISME J 8:830–840
Woodhams DC, Brandt H, Baumgartner S, Kielgast J, Küpfer E, Tobler U, Davis LR, Schmidt BR, Bel C, Hodel S, Knight R, McKenzie V (2014) Interacting symbionts and immunity in the amphibian skin mucosome predict disease risk and probiotic effectiveness. PLoS One 9, e96375
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522
Culp CE, Iii JOF, Belden LK (2007) Identification of the natubal bacterial microflora on the skin of eastern newts, bullfrog tadpoles and redback salamanders. Herpetologica 63:66–71
Fierer N, Hamady M, Lauber CL, Knight R (2008) The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A 105:17994–17999
Bolger D, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R (2010) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Price MN, Dehal PS, Arkin AP (2010) FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Statist 11:265–270
Faith DP, Baker AM (2006) Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinform Online 2:121–128
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Champaign
Bates D, Maechler M, Bolker BM, Walker SC (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48
Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R (2011) UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169–172
Rees GN, Baldwin DS, Watson GO, Perryman S, Nielsen DL (2004) Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Anton Leeuw Int J G 86:339–347
Okasen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoencs E, Wagner H (2016) Vegan: community ecology package. R package version 2.4-0
Hammer O, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:9
Longo AV, Savage AE, Hewson I, Zamudio KR (2015) Seasonal and ontogenetic variation of skin microbial communities and relationships to natural disease dynamics in declining amphibians. Roy Soc Open Sci 2:140377
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM (2014) Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642
Atlas RM (2010) Handbook of microbiological media. CRC Press, Taylor and Francis Group, Boca Raton
McKenzie VJ, Bowers RM, Fierer N, Knight R, Lauber CL (2012) Co-habiting amphibian species harbor unique skin bacterial communities in wild populations. ISME J 6:588–596
Bataille A, Lee-Cruz L, Tripathi B, Kim H, Waldman B (2016) Microbiome variation across amphibian skin regions: implications for chytridiomycosis mitigation efforts. Microb Ecol 71:221–232
Walke JB, Becker MH, Loftus SC, House LL, Cormier G, Jensen RV, Belden LK (2014) Amphibian skin may select for rare environmental microbes. ISME J 8:2207–2217
Crowhurst RS, Faries KM, Collantes J, Briggler JT, Koppelman JB, Eggert LS (2011) Genetic relationships of hellbenders in the Ozark highlands of Missouri and conservation implications for the Ozark subspecies (Cryptobranchus alleganiensis bishopi). Conserv Genet 12:637–646
Feist SM, Briggler JT, Koppelman JB, Eggert LS (2014) Within-river gene flow in the hellbender (Cryptobranchus alleganiensis) and implications for restorative release. Conserv Genet 15:953–966
Zilber-Rosenberg I, Rosenberg E (2008) Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32:723–735
Unger SD, Rhodes OE, Sutton TM, Williams RN (2013) Population genetics of the Eastern Hellbender (Cryptobranchus alleganiensis alleganiensis) across multiple spatial scales. PLoS One 8, e74180
Olson ZH, Burgmeier NG, Zollner PA, Williams RN (2013) Survival estimates for adult Eastern Hellbenders and their utility for conservation. J Herpetol 47:71–74
Bodinof CM, Briggler JT, Junge RE, Mong T, Beringer J, Wanner MD, Schuette CD, Ettling J, Millspaugh JJ (2012) Survival and body condition of captive-reared juvenile Ozark hellbenders (Cryptobranchus alleganiensis bishopi) following translocation to the wild. Copeia 2012:150–159
Becker MH, Richards-Zawacki CL, Gratwicke B, Belden LK (2014) The effect of captivity on the cutaneous bacterial community of the critically endangered Panamanian golden frog (Atelopus zeteki). Biol Conserv 176:199–206
Merrifield DL, Rodiles A (2015) The fish microbiome and its interactions with mucosal tissues. In: Mucosal health in aquaculture. Academic, Oxford, UK, pp 273–295
Pfingsten RA (1989) The status and distribution of the hellbender, Cryptobranchus alleganiensis, in Ohio. Ohio J Sci 89:3
Lauer A, Simon MA, Banning JL, Lam BA, Harris RN (2008) Diversity of cutaneous bacteria with antifungal activity isolated from female four-toed salamanders. ISME J 2:145–157
Michaels CJ, Antwis RE, Preziosi RF (2014) Impact of plant cover on fitness and behavioural traits of captive red-eyed tree frogs (Agalychnis callidryas). PLoS One 9(4):295207
Roth T, Foley J, Worth J, Piovia-Scott J, Pope K, Lawler S (2013) Bacterial flora on Cascades frogs in the Klamath mountains of California. Comp Immunol Microb 36:591–598
Lam BA, Walke JB, Vredenburg VT, Harris RN (2010) Proportion of individuals with anti-Batrachochytrium dendrobatidis skin bacteria is associated with population persistence in the frog Rana muscosa. Biol Conserv 143:529–531
Olson ME, Gard S, Brown M, Hampton R, Morck DW (1992) Flavobacterium indologenes infection in leopard frogs. JAVMA J Am Vet Med A 201:1766–1770
Taylor SK, Williams ES, Thorne ET, Mills KW, Withers DI, Pier AC (1999) Causes of mortality of the Wyoming toad. J Wildl Dis 35:49–57
Densmore CL, Green DE (2007) Diseases of amphibians. ILAR J 48:235–254
Geng Y, Wang KY, Zhou ZY, Li CW, Wang J, He M, Yin ZQ, Lai WM (2011) First report of a ranavirus associated with morbidity and mortality in farmed Chinese giant salamanders (Andrias davidianus). J Comp Pathol 145:95–102
Becker MH, Harris RN (2010) Cutaneous bacteria of the redback salamander prevent morbidity associated with a lethal disease. PLoS One 5, e10957
Bletz MC, Loudon AH, Becker MH, Bell SC, Woodhams DC, Minbiole KPC, Harris RN (2013) Mitigating amphibian chytridiomycosis with bioaugmentation: characteristics of effective probiotics and strategies for their selection and use. Ecol Lett 16:807–820
Acknowledgments
We would like to thank members of the Williams lab for assistance in revising this document. Special thanks also go to Jyothi Thimmapuram from the Purdue Bioinformatics Core, Phillip San Miguel and Paul Parker from the Purdue Genomics Core for assistance in project design, Bart Kraus for assistance in field collection, and Ardith Wang for assistance in sequencing preparation. Special thanks to the Missouri Department of Conservation for their interest and support of this project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
Funding for this study was provided by Purdue University.
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 187 kb)
Rights and permissions
About this article
Cite this article
Hernández-Gómez, O., Kimble, S.J.A., Briggler, J.T. et al. Characterization of the Cutaneous Bacterial Communities of Two Giant Salamander Subspecies. Microb Ecol 73, 445–454 (2017). https://doi.org/10.1007/s00248-016-0859-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0859-9